Introduction¶
Together with a cluster of genes called cas, CRISPR-Cas systems constitute a defence mechanism against mobile genetic elements, such as phages. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) elements consist in a succession of 24-47 base-pair long Direct Repeats (DR) separated by similarly sized unique sequences called spacers.
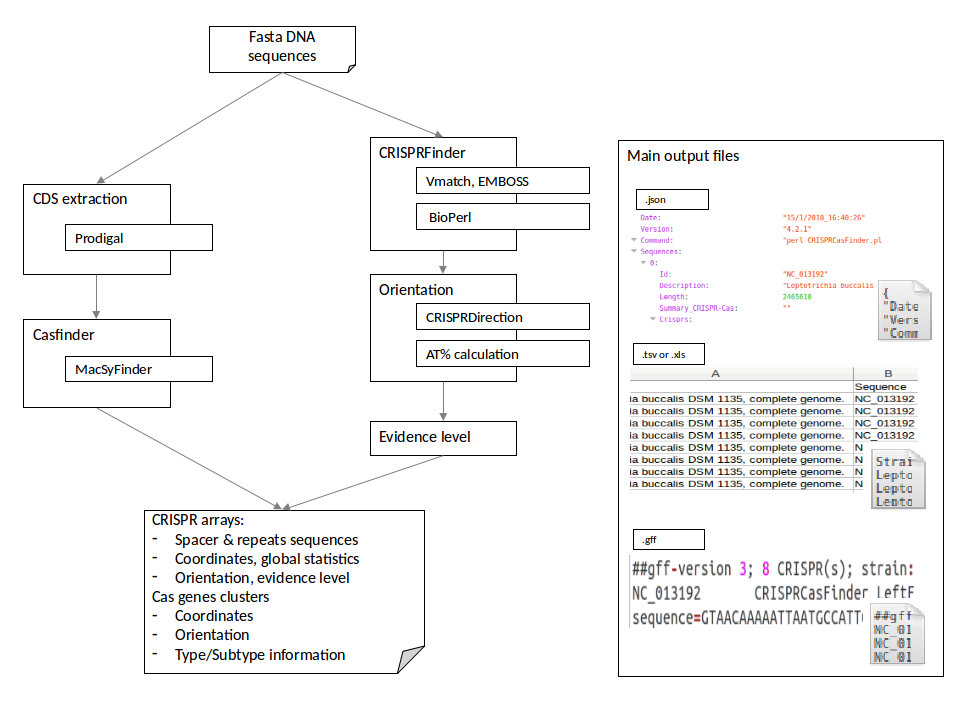
CRISPRCasFinder (a combination of CRISPRFinder version 4.2 and MacSyFinder version 2) allows the identification of both CRISPR arrays and cas genes, while providing new functions. CRISPRCasFinder is available at https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index.
- Current version: 4.2.5
- Release date: January 2018
- Download access of the stand-alone program (Linux or Mac OS): https://crisprcas.i2bc.paris-saclay.fr/Home/DownloadFile?filename=CRISPRCasFinder.zip
- Help concerning the installation of CRISPRCasFinder: attachment:"installer_README.txt"
CRISPRCasFinder is part of a project named "CRISPR-Cas++". This project was funded in part by Institut Français de Bioinformatique (IFB) grant ANR – 11 – INSB – 0013 ("CRISPRCas++").
A simplified workflow is shown below: